Address: Yuhangtang Road No.388, Hangzhou, China
Email: zhenhongdu[at]zju.edu.cn
Github
Biography: I’m currently a 4th year Ph.D. student at Zhejiang University, in a group managed by Prof. Ke Si and Prof. Wei Gong. Before joining ZJU, I obtained my B.E. from Hefei University of Technology, in June, 2020.
Interests:
1. Advanced imaging techniques
2. Deep learning in bio-imaging
3. Inverse problem
4. Computational Imaging
Education:
1. Ph.D. in Zhejiang University. (2020-)
2. B.E. in Hefei University of Technology. (2016-2020)
Hornors:
Excellent Graduate of Anhui Province (Bachelor’s degree) – (Anhui Province, 2020)
Excellent Graduate of Hefei University of Technology (Bachelor’s degree) – (Hefei University of Technology, 2020)
China National Scholarship of undergraduate (Top 1%) – (Hefei University of Technology, 2017)
Selected publications
-
Deep learning-assisted near-Earth asteroid tracking in astronomical images
Zhenhong Du, Hai Jiang, Xu Yang, Hao-Wen Cheng, and
Jing Liu
Advances in Space Research, 2024
, 73, 10, 5349-5362
The large number of near-Earth asteroids (NEAs) has greatly impacted human space activities and Earth security. However, detecting NEAs in astronomical images with complex, varying backgrounds is still extremely challenging. In this paper, we propose a deep segmentation assisted asteroid tracking algorithm, termed DSAT, to construct a possible pipeline for faint NEA tracking in astronomical images. First, the single-frame object detection problem is converted to a segmentation problem, enabling robust extraction of faint potential moving objects. Then, a multiframe motion prior-based moving object tracking algorithm is proposed to find real NEAs. We further propose a distance tolerance criterion to help DSAT achieve effective tracking in practical situations when detection has partially failed. Finally, the pipeline is tested with both simulated and real astronomical images at different SNRs and in crowded fields. The results showed that our pipeline has the potential to detect and track faint NEAs in complex backgrounds. Our code is publicly available at https://github.com/zhenhongdu/DeepSegAsteroidTracker.
-
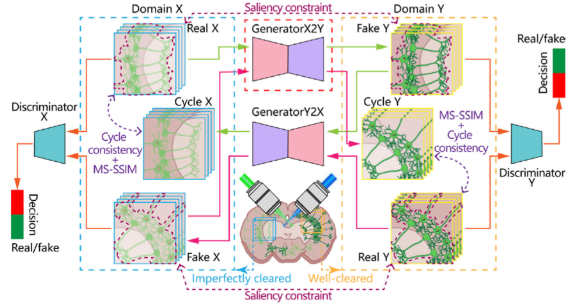
Three-Dimensional Virtual Optical Clearing With Cycle-Consistent Generative Adversarial Network
Jiajia Chen*, Zhenhong Du*, and
Ke Si
Frontiers in Physics, 2022
, 10, 965095
High-throughput deep tissue imaging and chemical tissue clearing protocols have brought out great promotion in biological research. However, due to uneven transparency introduced by tissue anisotropy in imperfectly cleared tissues, fluorescence imaging based on direct chemical tissue clearing still encounters great challenges, such as image blurring, low contrast, artifacts and so on. Here we reported a three-dimensional virtual optical clearing method based on unsupervised cycle-consistent generative adversarial network, termed 3D-VoCycleGAN, to digitally improve image quality and tissue transparency of biological samples. We demonstrated the good image deblurring and denoising capability of our method on imperfectly cleared mouse brain and kidney tissues. With 3D-VoCycleGAN prediction, the signal-to-background ratio (SBR) of images in imperfectly cleared brain tissue areas also showed above 40% improvement. Compared to other deconvolution methods, our method could evidently eliminate the tissue opaqueness and restore the image quality of the larger 3D images deep inside the imperfect cleared biological tissues with higher efficiency. And after virtually cleared, the transparency and clearing depth of mouse kidney tissues were increased by up to 30%. To our knowledge, it is the first interdisciplinary application of the CycleGAN deep learning model in the 3D fluorescence imaging and tissue clearing fields, promoting the development of high-throughput volumetric fluorescence imaging and deep learning techniques.
-
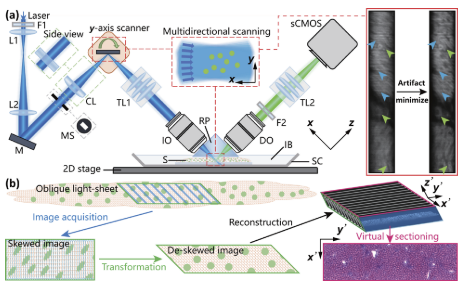
Ultrafast 3D histological imaging based on a minutes-time scale tissue clearing and multidirectional selective plane illumination microscopy
Jiajia Chen, Zhenhong Du, Chenhao Xu, Xiao Xiao, Wei Gong, and
Ke Si
Optics Letters, 2022
, 47, 17, 4331–4334
Conventional histopathological examinations are time-consuming and labor-intensive, and are insufficient to depict 3D pathological features intuitively. Here we report an ultrafast 3D histological imaging scheme based on optimized selective plane illumination microscopy (mSPIM), a minutes-time scale clearing method (FOCM), and a deep learning-based image enhancement algorithm (SRACNet) to realize histological preparation and imaging of clinical tissues. Our scheme enables 1-minute clearing and fast imaging (up to 900 mm2/min) of 200 \textmum-thick mouse kidney slices at micron-level resolution. With hematoxylin and eosin analog, we demonstrated the detailed 3D morphological connections between glomeruli and the surrounding tubules, which is difficult to identify in conventional 2D histology. Further, by the preliminary verification on human kidney tissues, this study will provide new, to the best of our knowledge, feasible histological solutions and inspirations in future 3D digital pathology. \copyright 2022 Optical Society of America


